Projekte
Overview given by Prof. Markus List
DROP2AI stands for Drug Response Prediction using Proteomics and AI.
The BMBF-funded DROP2AI project aims to predict drug response from transcriptomics and proteomics drug screens using state of the art AI methods. We will leverage data from several in-house cellular drug response screens involving hundreds of approved drugs and hundreds of cell lines as well as proteome expression data collected for the same systems to test various ML and AI strategies for drug response prediction. The resulting prediction models will be tested in silico using publicly available data, experimentally verified via focused laboratory experiments in cell lines, organoids and mouse models and applied to data collected for cancer patients in the NCT/DKTK-MASTER (Molecularly Aided Stratification for Tumor Eradication) trial. By covering the whole chain from data curation, method development towards experimental and clinical application as well as making all data and tools available as part of the existing ProteomicsDB platform, we will help in advancing systems medicine in general, and the work of molecular tumor boards in particular.
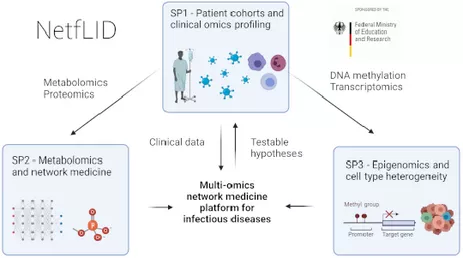
In the DFG-funded project DyHealthNet we build an explorative analysis platform for population cohorts. This work is based on the CHRIS cohort data and carried out in collaboration with the Friedrich-Alexander-Universität Erlangen, the Eurac Research Center und the University of Bolzano in South Tyrol (Italy).
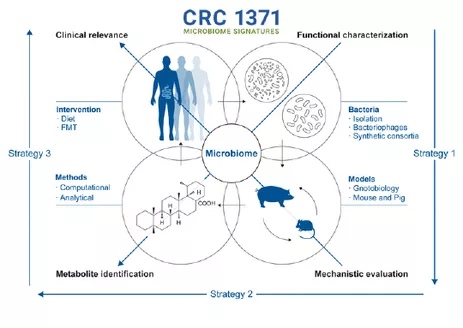
The DFG-funded Collaborative Research Center 1371 for Microbiome Signatures investigates the functional relevance of the microbiome in the digestive tract. The goal is to determine their precise contribution in a disease-specific manner. SSMS is the abbreviation of “shallow shotgun metagenomics sequencing”.
DYS-SPONGE
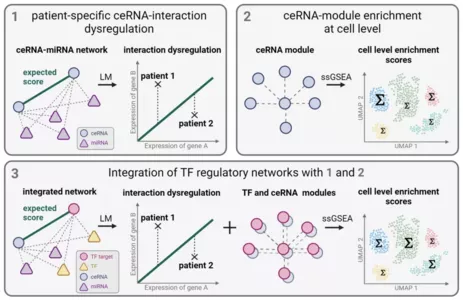
A joint framework integrating microRNA and transcription factor regulation analysis at bulk and single-cell level: Within DYS-SPONGE, we study the complementary effects of transcription factors, micro RNAs and competing endogenous RNAs in gene regulation. This project is funded by Linde/Munich Data Science Institute.
CoBiNet

CoBiNet develops methods to retrieve context-specific and bias-reduced high-resolution protein-protein interaction networks. This research project is enabled by the Klaus Tschira Foundation and a collaboration with FAU (Biomedical Network Science Lab) in Erlangen and IEO (Computational Cancer Biology Lab) in Milano.