Accurate protein function prediction for plant proteins - Benchmarking and extending DeepGOPlus
Topic
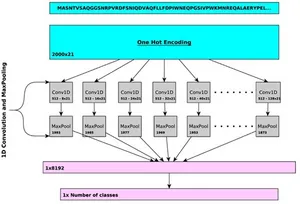
This project focuses on enhancing protein function prediction using deep learning, specifically by building upon the DeepGOPlus model. DeepGOPlus is a method that combines deep convolutional neural networks with sequence similarity for protein function prediction.
Participants will engage in various tasks, including:
- Data Expansion: Identifying and incorporating additional training data, particularly focusing on plant proteins.
- Model Refinement: Retraining or fine-tuning DeepGOPlus with the newly acquired data to improve its predictive performance for plant proteins.
- Feature Optimization: Enhancing the model by integrating new features, such as inputs from PSI-BLAST, Hidden Markov Models (HMMs), or structural information from models like ESM by Meta.
- Performance Evaluation: Assessing the model's performance using standards established by the Critical Assessment of Functional Annotation (CAFA) challenge.
- Comparative Analysis: Comparing the enhanced model's performance with existing approaches like InterPro or PSI-BLAST.
- Application: Predicting Gene Ontology (GO) terms for plant proteins within the "The Proteomes that Feed the World" project by the Elite Network of Bavaria.
Aim
The primary objective is to improve the accuracy and applicability of protein function prediction models for plant proteins by:
- Expanding Training Data: Sourcing and integrating additional plant-specific protein data to enhance model training.
- Model Enhancement: Fine-tuning DeepGOPlus with the new data and incorporating additional features to boost predictive performance. Optionally, if interested and time allows, we would integrate the structural information of the proteins as an input to the model. One suggestion is the protein embeddings via the ESM model from Meta.
- Rigorous Evaluation: Employing CAFA challenge standards to evaluate and validate the model's performance.
- Practical Application: Applying the refined model to annotate plant proteins in "The Proteomes that Feed the World" project.
- Benchmarking: Optionally, comparing the model's predictions with those from established tools like InterPro or PSI-BLAST to assess relative performance.
General Schedule
Phase 1: Methods, Tools, Techniques
The first phase consists of a series of seminars in which you will learn the basics of various topics necessary for the project. The seminars will be a mix of presentations by team members of our research group, practical sessions where applicable, and short presentations prepared by the participants:
- Kickoff Seminar: In the first seminar, we will discuss the organizational part and provide you with insights into the project. We will demonstrate a high-level overview of our workflows to enable you to select your focus area, scope the project work, and later on deliver a successful project.
- Protein function prediction: Seminar covering the definition of protein function and the existing methods for their prediction. This covers topics such as Gene Ontologies and classic and novel GO term prediction methods.
- Machine Learning: This lecture will cover the machine learning techniques we apply. Mainly deep learning, fine-tuning, and interpretation. You will get insights into the DeepGOPlus model, the different possibilities for extensions, access to our infrastructure, and how we use it.
- Experimentation best practices: This will be organized in a block session covering experimentation best practices from our experience with deep learning, software development, and will touch on topics such as implementing modules, testing several components, iterating fast, etc.
Additional Topics: In case you want to get deeper knowledge we are open to holding an additional seminar with a topic of your choice. You decide.
Phase 2: Research project planning
In the second phase, we want you to prepare a detailed project plan. At the end of this phase, you will present your plan and discuss it with us. We will assist you during the planning of your project and provide you with feedback to ensure that you are able to bring your project to a success. Most importantly, you should discuss the following points:
- Focus Area Selection: You, as a group, will select and agree on a strategy to approach the project. Based on that, we will provide you with resources and alternatives to consider for scoping and planning your project.
- Requirement Analysis: You, as a group, will have to discuss and plan what exact research questions proposed by us (or by you) will be covered and by whom. This will include defining the research question, data needs, and technical requirements for the chosen tasks.
- Project Organization: Projects work best if they are well organized. In your plan, we expect to see clear milestones, a time plan, and a reasonable task distribution among individual team members. If you like, you could use tools like Monday.com to organize your work. Communication in our lab is conducted via Slack.
Frameworks and Languages: We use the common Python stack for numerical computing (numpy, pandas) along with TensorFlow/Keras for deep learning. In selected cases, we also use R for plotting and/or analyses.
Phase 3: Implementation and Research
This is the main phase of your project. According to your plan, you will implement, integrate, and test your work according to the plan. We will hold weekly to discuss your progress.
- Semester Work: We would like you to work during your semester. You do not need to come in person all the time although we provide a large room where you can work. However, we encourage you to come to our research group. If you are on site, we will be able to assist and guide you more directly. Virtually, we can also offer office-hours for addressing questions.
- Full-Time Block: Depending on how you plan your project and your progress we can have a two to three-week-long intensive block during or at the end of your semester. The specific time and requirements will be discussed with you.
- Submission: At the end of this project, you will need to provide some deliverables that include research questions answered, features implemented, code documentation, and testing. Ideally, this would evolve as your implementations progress during the semester. We expect you to write a report and give a presentation of your work in our research group.
Skills Gained
By participating in this project, you will improve your programming skills, gain knowledge in the field of protein function prediction, and gain experience in deep learning. You will also gain a solid basis in software engineering and development that will be very helpful for your career, both in academia and in the industry. Participants will develop:
- Advanced Programming Skills: Enhanced proficiency in Python, particularly in deep learning frameworks.
- Bioinformatics Expertise: Understanding of protein function prediction and related bioinformatics tools.
- Data Analysis Skills: Experience in data sourcing, preprocessing, and integration for model training.
- Model Development and Evaluation: Skills in training, fine-tuning, and assessing deep learning models using industry standards.
- Research Application: Experience in applying computational models to real-world biological data.
Organisation
Programming Language: Python
Must-have Skills:
- Intermediate Python programming
- Basic understanding of deep learning concepts
- Interest in applied research and writing good-quality code
- Interest in bioinformatics and protein function prediction
Good-to-have Skills:
- Experience with deep learning frameworks (e.g., TensorFlow, PyTorch)
- Knowledge of common and state-of-the-art deep learning architectures (Transformers, RNNs, CNNs)
- Familiarity with bioinformatics tools (e.g., PSI-BLAST, InterPro)
- Knowledge of protein structure prediction models (e.g., ESM by Meta)
- Basic skills in Git and GitHub
Supervisors:
- Armin Soleymaniniya - primary - Bioinformatics specialist
- Joel Lapin - primary - Deep learning specialist
- Mathias Wilhelm - secondary - Overall guidance
Grading:
- Presentation [max. 30 minutes, whole team]
- Report [~10 pages, including all sections]
Team Size: 3 participants
Submission:
- Git repository with code and documentation
- A comprehensive report detailing the project
Location/Rooms: We are very flexible with regard to the time slots (and location to some degree). As of right now, we expect phases 1-3 will take place either in Freising/Weihenstephan (or via Zoom – but this is not preferred at all). Lecture preferably in the afternoon (tentative Thursday 16-18), project meetings preferably at the same time. However, we are flexible with respect to the day and will decide with your input. We also have a student room that you can use but you may also work from home if you so desire. We would like to welcome you to our institute. The full-time block will take place - if required - in Freising/Weihenstephan. The exact weeks will be decided upon once we know if it is required. Online participation is possible for all parts except the full-time block.
Material
All materials are made available in TUM Moodle.
Literature
GitHub repos of relevance:
Relevant publications:
- DeepGOPlus: improved protein function prediction from sequence
- DeepGO: predicting protein functions from sequence and interactions using a deep ontology-aware classifier
- The CAFA Challenge
- The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens
- Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences