Tackling class imbalance in mass spectrometry-based peptide property prediction
Topic
Proteomics, the large-scale study of proteins, is essential for understanding biological processes, disease mechanisms, and biomarker discovery. Mass spectrometry (MS) is a cornerstone technology in proteomics, providing high-throughput analysis of peptides and proteins. Accurate prediction of peptide properties using deep learning, such as charge state, is crucial for improving MS data interpretation, enabling more reliable peptide spectrum prediction, and enhancing both identification and quantification of proteins.
However, the challenge of class imbalance, where certain charge states are underrepresented in the data, often hampers the performance of machine learning models in this domain. Standard predictive models struggle to be accurate across all classes, leading to biased outcomes and sub-optimal charge state distribution representations. Tackling this issue requires innovative methods to address class imbalance and ensure robust performance for all charge states.
This project focuses on developing and evaluating approaches to mitigate class imbalance in charge state prediction. You will explore advanced techniques to improve predictive performance, aiming to set the foundation for better peptide property prediction models that enhance MS-based proteomics workflows.

Aim
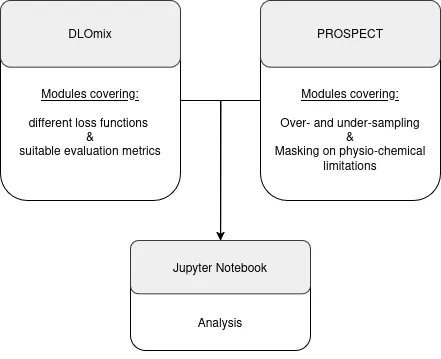
You will work on a larger project that can be divided into sub-projects tailored to your preferences and skills. The overarching goal is to address class imbalance issues by modularly extending the existing DLOmix and PROSPECT repositories.
The framework you develop will leverage functions from DLOmix and PROSPECT to enable modular testing and evaluation of both individual and combined mitigation techniques. The modules you will create include:
- Model-based techniques:
- Different loss functions (e.g., weighted losses, combinations of losses).
- Suitable evaluation metrics for robust performance assessment.
- Data processing techniques:
- Over- and under-sampling strategies to balance datasets.
- Masking based on physio-chemical limitations
Optional tasks (if time permits):
- Investigating the effects of post-translational modifications on predictions.
- Analyzing the impact of rare training sequences.
- Identifying and mitigating experimental batch effects.
General Schedule
Phase 1: Methods, Tools, Techniques
The first phase consists of a series of seminars and lectures in which you will learn the basics of various topics necessary for the project. There will be a mix of presentations by team members of our research group, practical sessions where applicable, and short presentations prepared by the participants:
Kickoff Seminar - In the first seminar, we will discuss the organizational part and provide you with insights into the project. We will demonstrate a high-level overview of our workflows, the scope of the project work, and our expectations with regard to the success of the project.
Lecture 1 (Mass spectrometry and charge states) - In this lecture, you will learn about the experimental and technical backgrounds of mass spectrometry and how all this influences the charge state distribution of peptides.
Lecture 2 (Deep Learning, Predicting Charge state distributions) - The second lecture focuses on state-of-the-art deep learning approaches to predict charge state distributions. Further, there will be an introduction to the DLOmix, a Python framework for Deep Learning in Proteomics, and PROSPECT (PROteometools SPECTrum compendium) large annotated datasets. You will use both of these during your project.
Lecture 3 (Working with git as a team) - The third lecture will provide you with a project management system for working on larger coding projects. We will cover the concepts of issues, branches, pull requests, and the review process.
Lecture 4 (Sub-problems of the project) - In the final lecture, we delve into the sub-problems of the project, and you will start planning how to tackle them.
Additional Topics - In case you want to get deeper knowledge, we are open to holding an additional seminar on a topic of your choice. You decide.
Phase 2: Research project planning
In the second phase, we want you to prepare a detailed project plan. At the end of this phase, you will present your plan and discuss it with us. We will assist you during the planning of your project and provide you with feedback to ensure that you can bring your project to a success. Most importantly, you should discuss the following points:
Focus Areas Selection - As a group, you will select and agree on a strategy to integrate the new functionalities. Based on that, we will provide you with resources and alternatives
for scoping and planning your project.
Task specification and allocation - As a group, you will have to discuss and plan what exact modules and associated research questions will be covered and by whom. This will include a requirement analysis for each module and a detailed timeline. We advise you to use project management software like Notion or Jira.
Frameworks and Languages - We use the common Python stack for numerical computing (numpy, pandas) along with TensorFlow/Keras and maybe PyTorch for deep learning.
Phase 3: Implementation and Research
This is the main phase of your project. According to your plan, you will implement, integrate, and test your work with our existing architecture. We will hold group weeklies to discuss your individual and overall progress.
Semester Work - We would like you to work during your semester. You do not need to come to Freising in person, but if you would like to work on-site, we provide a large office.
Full-Time Block - Depending on your progress according to your plan, there will be a one to three-week-long intensive block at the beginning of the semester break. The specific time will be discussed with you.
Submission - At the end of this project, you will need to provide some deliverables that include answers to research questions, features implemented, code documentation, and testing. This will evolve as your implementations progress during the semester. Additionally you will write a report and present your work in our research group seminar.
Skills Gained
By participating in this project, you will improve your programming skills and gain experience in data analysis for proteomics, deep learning, and software engineering. All of these will be very helpful for your career in academia and industry.
Organisation
Programming language: Python
Must-have skills
- Intermediate programming skills in Python
- Basic understanding of deep learning concepts
- Interest in proteomics
Good-to-have skills
- Knowledge of good software engineering practices
- Knowledge of TensorFlow/Keras, PyTorch, and Hugging Face
- Basic skills in Git and GitHub
Supervisors
- Jesse Angelis - primary
- Ayla Schröder - primary
- Mathias Wilhelm - secondary
Grading Based on individual contribution in weekly meetings, code issues/commits/pull requests/documentation, final presentation, and report.
Team size This project is designed for a team of 4 people.
Location OG-L 19, Maximus-von-Imhof-Forum 3, 85354 Freising
Dates We are flexible to some extent in regard to the time slots. We tentatively scheduled this as follows:
- 24.04.25 (4 pm - 6 pm) - Kickoff Seminar and Lecture 1 - On-Site
- 08.05.25 (4 pm - 6 pm) - Lecture 2 & 3 - On-Site
- 15.05.25 (4 pm - 6 pm) - Lecture 4 & Beginning project plan - On-Site
- 22.05.25 - 24.07.25 (4 pm - 5 pm) - Weekly group meetings - Online or On-Site
- Intensive Block (1-3 weeks, earliest 04.08.25) - On-Site
Material
All materials are made available in TUM Moodle.
Literature
GitHub repositories of relevance:
- DLOmix framework
- PROSPECT training data
- https://huggingface.co/Wilhelmlab
Literature:
- CPred publication (https://doi-org.eaccess.tum.edu/10.1021/acs.analchem.4c01107)
- Guan et al. (10.1074/mcp.TIR119.001412)
- Gorshkov et al. (https://doi-org.eaccess.tum.edu/10.1021/acs.analchem.3c04270)