Identification of microbes providing colonization resistance against enteric infections
Work within the “Translational GI Microbiome Research Platform” funded by the German Center of Infection Research (DZIF) elucidates principles of microbiota-based intervention with virulence of intestinal pathogens (focus: EHEC and Salmonella). In the past, we identified different commensal bacteria that protect against enteric S. Tm infection using comparative microbiome analysis. The gnotobiotic OMM12 mouse model enabled us to demonstrate causality in protection and to uncover on the underlying mechanisms.
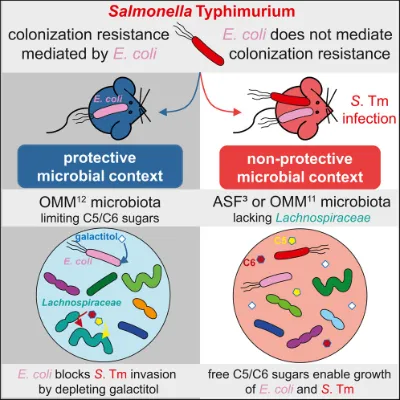
Commensal E. coli can block S. Tm colonization, a mechanism that is microbial context dependent (Eberl et al., 2021). In an infection-permissive context (e.g. mice colonized with a low-diverse microbiota), a high diversity of carbon sources is available which enables growth of both, E. coli and S. Tm. In mice that were stably colonized with OMM12, establishing a protective context, E. coli depleted galactitol, a substrate otherwise fueling S. Tm colonization. Here, Lachnospiraceae, capable of consuming C5 and C6 sugars, critically contributed to colonization resistance. We propose that E. coli provides colonization resistance by depleting a limited carbon source when in a microbial community adept at removing simple sugars from the intestine.
Additionally, using a murine Salmonella infection model, we found that microbiota alterations and enrichment of Mucispirillum spp. correlated with protection against infection. This genus belongs to the phylum Deferribacteres and is a prevalent but low abundant member of the rodent, pig and human microbiota (Herp et al., 2021). Mucispirillum spp. is enriched under intestinal inflammatory conditions (Loy et al., 2017) and has recently also been causally linked to the development of Crohn’s disease-like colitis in immunodeficient mice (Herp et al., 2021). Using the OMM12 mouse model, we could show that Mucispirillum schaedleri also protects against S. Tm colitis by interfering with invasion gene expression and by competing for anaerobic electron acceptors like nitrate. Our study establishes M. schaedleri, a core member of the murine gut microbiota, as key antagonist of S. Tm virulence in the gut (Herp et al., 2019).
Publications:
Eberl, C., Weiss, A.S., Jochum, L.M., Durai Raj, A.C., Ring, D., Hussain, S., Herp, S., Meng, C., Kleigrewe, K., Gigl, M., et al. (2021). E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe. 29(11), 1680-1692 e1687. DOI:10.1016/j.chom.2021.09.004
Herp, S., Brugiroux, S., Garzetti, D., Ring, D., Jochum, L.M., Beutler, M., Eberl, C., Hussain, S., Walter, S., Gerlach, R.G., et al. (2019). Mucispirillum schaedleri Antagonizes Salmonella Virulence to Protect Mice against Colitis. Cell Host Microbe. Published online 2019/04/23 DOI: 10.1016/j.chom.2019.03.004.
Herp, S., Durai Raj, A.C., Salvado Silva, M., Woelfel, S., and Stecher, B. (2021). The human symbiont Mucispirillum schaedleri: causality in health and disease. Med Microbiol Immunol. 210(4), 173-179. DOI: 10.1007/s00430-021-00702-9.
Loy, A., Pfann, C., Steinberger, M., Hanson, B., Herp, S., Brugiroux, S., Gomes Neto, J.C., Boekschoten, M.V., Schwab, C., Urich, T., et al. (2017). Lifestyle and Horizontal Gene Transfer-Mediated Evolution of Mucispirillum schaedleri, a Core Member of the Murine Gut Microbiota. mSystems. 2(1). DOI: 10.1128/mSystems.00171-16.
External links:
https://www.dzif.de/en/working-group/translational-microbiome-research-platform
https://www.dzif.de/en/working-group/cegimir-centre-gi-microbiome-research
https://www.dzif.de/de/auf-der-suche-nach-bakteriencocktails-zur-bekaempfung-von-infektionen
https://www.dzif.de/de/salmonellen-infektion-schuetzende-darmbakterien-identifiziert
