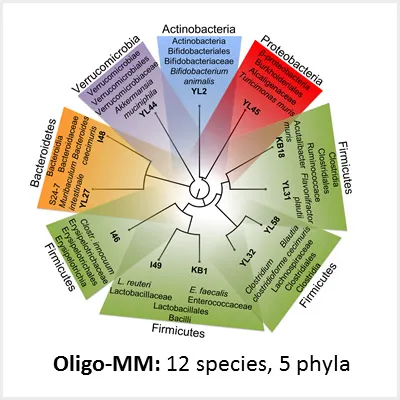
OMM12: A synthetic bacterial community
A central challenge in gut microbiome research is to understand how interactions between the individual microorganisms affect community assembly, dynamics and functionality. Gnotobiotic mice colonized with defined microbial communities have become essential tools to address this question. While a majority of studies uses human-derived bacteria, we have developed a model community consisting of twelve phylogenetically diverse bacteria isolated from mice (Brugiroux et al., 2016) (Fig. 3). In addition, we have developed a comprehensive set of protocols and analysis tools, allowing us to study this community in its native murine host. This Oligo-Mouse-Microbiota (OMM12) is unique because it recapitulates central physiologic and functional traits of a complex mouse microbiome. Furthermore, the OMM12 exhibits long-term stability in gnotobiotic mice and its composition is reproducible among different animal facilities. Hence, this model is used by an increasing number of research groups world-wide.
To provide an ecological context, we extensively studied the interaction network and metabolic capabilities of this synthetic bacterial community, both of which are factors that determine community assembly, dynamics and functionality (Weiss et al., ISME 2021). We employed a bottom-up approach connecting outcomes of mono- and pairwise co-culture experiments with observations from complex communities in in vitro batch culture. Furthermore, we combined metabolomics analysis of spent culture supernatants with genome-informed pathway reconstruction and generated metabolic models of the OMM12 consortium. Together, this work identified key interaction patterns among OMM12 strains relevant in community assembly and functionality.
Publikations:
Brugiroux, S., Beutler, M., Pfann, C., Garzetti, D., Ruscheweyh, H.J., Ring, D., Diehl, M., Herp, S., Lotscher, Y., Hussain, S., et al. (2016). Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol. 2, 16215. DOI: 10.1038/nmicrobiol.2016.215.
Eberl C, Ring D, Münch PC, Beutler M, Basic M, Slack EC, Schwarzer M, Srutkova D, Lange A, Frick JS, Bleich A, Stecher B (2020). Reproducible Colonization of Germ-Free Mice With the Oligo-Mouse-Microbiota in Different Animal Facilities. Front Microbiol. DOI: 10.3389/fmicb.2019.02999
Weiss, A.S., Burrichter, A., Durai Raj A.C., von Strempel A., Meng, C., Kleigrewe K., Münch P.C., Rössler L., Huber C., Eisenreich W., Jochum L.M., Göing S., Jung K., Lincetto C., Hübner J., Marinos G., Zimmermann, J., Kaleta C., Sanchez A., Stecher B (2021). In vitro interaction network of a synthetic gut bacterial community. ISME . DOI: 10.1038/s41396-021-01153-z