Nucleic Acid and Bioanalytical Chemistry
Life originates from a set of rules shared by all organisms and encoded within genetic material, both DNA and RNA. As a result, much of modern biological research relies on modern approaches to efficiently read from- and write information into these molecules. In addition to DNA and RNA, nucleic acid chemistry now encompasses the synthesis, characterization and application of novel molecular constructs that extend or complement the functionality of the natural molecules. These xenobiotic nucleic acids (XNA) include modifications to the sugar, the nucleobase, or the backbone that modulate properties such as nuclease or thermal stability, structure, information storage capacity, or pharmacokinetics.
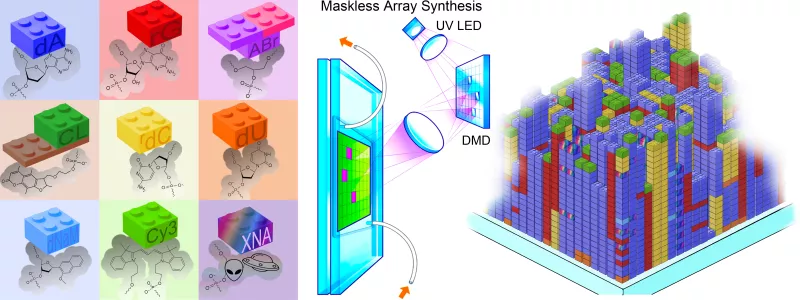
Our research group is particularly interested in the use of large-scale approaches to the synthesis, characterization and development of bioanalytical methods based on natural and xenobiotic nucleic acids. The scale of the approach is based on the use of photolithographic techniques adapted from the semiconductor industry. Nucleic acid photolithography uses an ultraviolet optical system to image the pattern displayed on a Digital Micromirror Device to a substrate where the light is used to selectively remove photolabile protecting groups from monomers of nucleic acids, which allows their sequence-defined polymerization on a massive scale. Synthesized in a few hours, these arrays hold from hundreds of thousands to a few million unique nucleic acid oligomers, including DNA, RNA or XNA, or combinations thereof. The arrays are then used to trap and quantify the abundance of DNA or RNA from cells or tissues for genomics or transcriptomics experiments, or to understand the molecular behavior of nucleic acids, including binding or enzymatic interaction with proteins.
Selected Publications:
Philipp L. Antkowiak, Jory Lietard, Mohammad Zalbagi Darestani, Mark M. Somoza, Wendelin J. Stark, Reinhard Heckel, Robert N. Grass (2020). Low cost DNA data storage using photolithographic synthesis and advanced information reconstruction and error correction. Nature Communications 11, 5345. doi.org/10.1038/s41467-020-19148-3
E. Schaudy, M. M. Somoza, J. Lietard (2020) L‐DNA Duplex Formation as a Bioorthogonal Information Channel in Nucleic Acid‐Based Surface Patterning. Chemistry: A European Journal. doi.org/10.1002/chem.202001871
V. Stoeger, A.-K. Holik, K. Hölz, T. Dingjan, J. Hans, J. P. Ley, G. E. Krammer, M. Y. Niv, M. M. Somoza, V. Somoza (2020) The bitter tasting amino acids L arginine and L isoleucine differentially regulate proton secretion via T2R1 signalling in human parietal cells in culture. Journal of Agricultural and Food Chemistry. doi.org/10.1021/acs.jafc.9b06285.
J. Lietard, M. Damha, M. M. Somoza (2019) Large-scale photolithographic synthesis of chimeric DNA/RNA hairpin microarrays to explore sequence specificity landscapes of RNase HII cleavage. Biochemistry 58(44), 4389-4397. doi.org/10.1021/acs.biochem.9b00806
J. Hans, J. Hoi, M. Pignitter, B. Lieder, J. Ley, J. Lietard, K. Hölz, M. M. Somoza, V. Somoza (2019) Identification of cinnamaldehyde as most effective fatty acid uptake reducing cinnamon-derived compound in differentiated Caco-2 cells compared to its structural analogs cinnamyl alcohol, cinnamic acid and cinnamyl isobutyrate. Journal of Agricultural and Food Chemis