From metabolite profiling to biomarker discovery
In order to understand the diversity of life at different molecular levels, nucleic acids, proteins, and metabolites are used as valuable analytical reporters to evaluate the status of a biological system. Although in principle encoded in the genome, proteins and metabolites add tremendous diversity to biological systems as the vast majority of biological processes are executed and controlled by the concerted action of proteins and/or metabolites. Among the molecules of life, the complex pattern of low-molecular weight metabolites reflects the various functional activities, transient effects, as well as endpoints of biological processes determined by the sum of its genetic features, regulation of gene and protein expression as well as dietary and environmental influences.
As the metabolome cannot be computed from the genome, the goal of metabolomics is to monitor and quantify low molecular weight molecules, produced by active cells under different conditions and times in their life cycles. Due to the enormous analytical progress made in recent years, metabolite profiling by means of state-of-the-art liquid chromatography - mass spectrometry (HILIC-MS/MS, UPLC-TOF-MS) reached a high level of sensitivity, increasing metabolite coverage, and a high sample throughput. As metabolomics is a data-driven technology, the transformation of raw data into information suitable for their biological interpretation is achieved by the implementation of sophisticated bioinformatics tools adapted to the field of research.
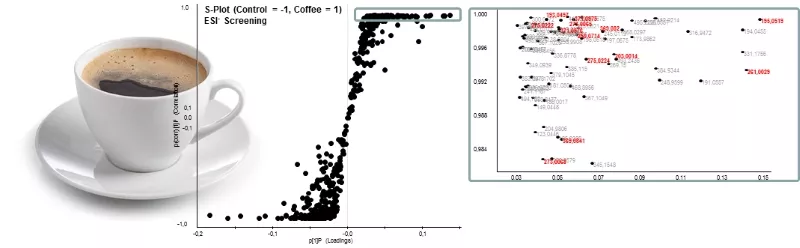
For the identification of key metabolites of bioactive constituents such as, e.g. alkaloids and polyphenols from coffee, circulating in the vascular system after food consumption, human intervention studies are conducted in cooperation with the ZIEL – Institute for Food and Health. The results of these help localize qualitative and quantitative differences in the metabolome of urine and blood-sample from coffee- and control group, respectively, by means of UPLC-ToF-MS (Fig. 1), followed by unequivocal identification of localized markers via exact mass, data base search, and subsequent chemical synthesis.
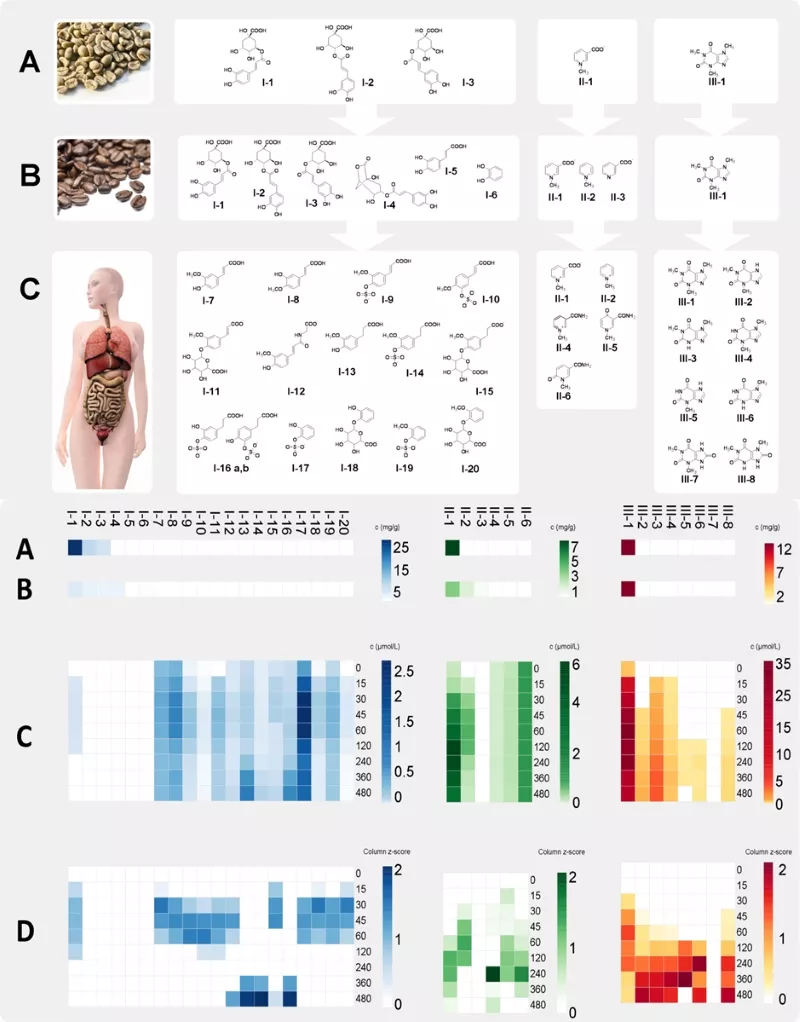
Stable isotope dilution analysis (SIDA)-based targeted UPLC-MS/MS methods are developed and applied for quantification and construction of pharmacokinetic profiles (Fig. 2). It became evident that the human organism quickly degraded green coffee derived chlorogenic acids (lower scheme, A) and their pyrolysis products formed during the roasting process (B) into phenylpropenic acids and phenylpropanic acids after consumption of roast coffee brew. Subsequently, these are conjugated into sulfate-, glycineamide- and glucuronic acid derivatives during phase II metabolism (C).
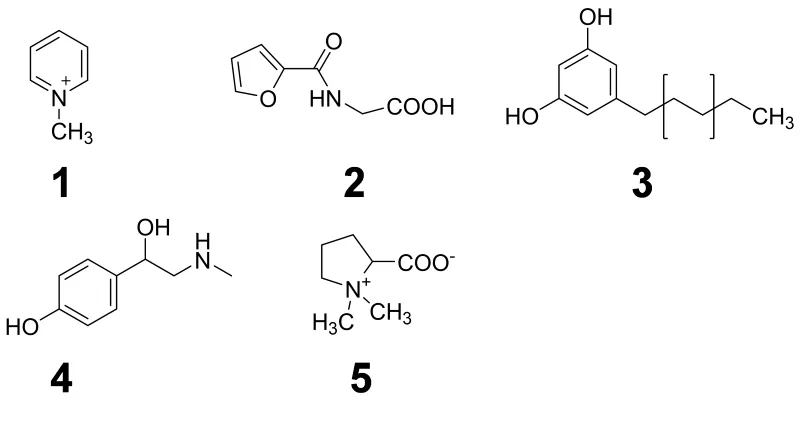
N-Methylpyridinium (II-2) ions originating from trigonelline during the coffee roasting process were identified as a coffee-specific biomarker in human plasma, saliva and urine. As scientists involved in health related dietary intervention studies often struggle with the accuracy of the study participants’ answers on frequency and quantity of consumed foods given in dietary questionnaires, the targeted HTP-quantitation of N-methylpyridinium in human urine by UPLC-SIDA-MS/MS has been applied as an objective tool for compliance control in human intervention studies on the health effects of coffee consumption.
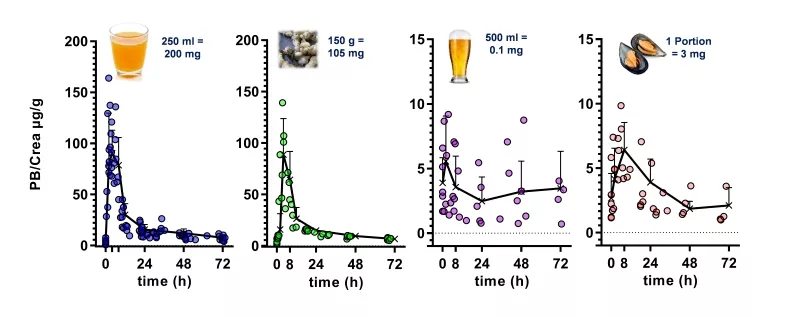
Such “dietary biomarkers” are valuable tools to check for a recommended abstinence of distinct foods and beverages in the wash-out phase of an intervention study as well as to validate the defined consumption of a distinct food under investigation in the intervention phase according to the study protocol. “Dietary biomarkers” (Fig. 3) such as, e.g. N-methylpyridinium cation (1) and furoyl-N-glycine (2) identified in urine after roast coffee consumption, alkylresorcinols (3) identified in urine after ingestion of wheat whole grains, synephrine (4) identified in urine after consumption of oranges and mandarins, and proline betaine (5) found in urine after consumption of citrus fruits and Chinese artichoke (Fig. 4), respectively, can contribute, therefore, to the quality of data sets, as participants deviating from the dietary protocol can be identified and the respective data excluded from further processing and interpretation.
[Translate to English:]
Current research is focusing on the discovery of specific dietary biomarkers for a broad range of foods and beverages and the development of a multiparametric high-throughput LC-MS method for a one-shot quantitation of a series of dietary biomarkers in urine.
SELECTED PUBLICATIONS:
Bader, M.; Lang, T.; Lang, R.; Hofmann, T. (2017) Synephrine as a Specific Marker for Orange Consumption. J. Agric. Food Chem.; DOI: 10.1021/acs.jafc.7b01941.
Lang, R.; Lang, T.; Bader, M.; Beusch, A.; Schlagbauer, V.; Hofmann, T. (2017) High-Throughput Quantitation of Prolilne Betaine in Foods and Suitability as a valid biomarker for citrus consumption. J. Agric. Food Chem., 65, 1613-1619.
Lang, R.; Dieminger, N.; Beusch, A.; Lee, Y.-M.; Dunkel, A.; Suess, B.; Skurk, T.; Wahl, A.; Hauner, H.; Hofmann, T. (2013) Bioappearance and pharmacokinetics of bioactives upon coffee consumption. Anal. Bioanal. Chem., 405, 8487 – 8503.
Krug, S.; Kastenmüller, G.; Stückler, F.; Rist, M.J.; Skurk, T.; Sailer, M.; Raffler, J.; Römisch-Margl, W.; Adamski, J.; Prehn, C.; Frank, T.; Engel, K.H.; Hofmann, T.; Luy, B.; Zimmermann, R.; Moritz, F.; Schmitt-Kopplin, P.; Krumsiek, J.; Kremer, W.; Huber, F.; Oeh, U.; Theis, F.J.; Szymczak, W.; Hauner, H.; Suhre, K.; Daniel, H (2012) The dynamic range of the human metabolome revealed by challenges. FASEB J., 26, 2607-2619.
Lang, R.; Wahl, A.; Stark, T.; Hofmann, T. (2011) Urinary N-methylpyridinium and trigonelline as candidate dietary biomarkers of coffee consumption. Mol. Nutr. Food Res., 55, 1613-1623.
