Nuclear Magnetic Resonance Spectroscopy (NMR)
NMR spectroscopy, a key spectroscopic tool in modern natural product chemistry, is used to perform 1-dimensional (1H, 13C, Dept-135) and 2-dimensional experiments (COSY, TOCSY, NOESY, ROESY, HSQC, HMBC) in order to elucidate the detailed chemical structure of a given target molecule.
In addition, quantitative NMR spectroscopic (qNMR) methods are developed, validated and applied to precisely quantitate organic molecules in complex mixtures such as, e.g. urine, plasma, food extracts, and to assess the purity of isolated molecules to be used as internal standards for quantitative studies or as reference material for bioactivity testing.
NMR spectrometer:
| AV 500 | |
|---|---|
| Magnet: | 500 MHz ultrashield plus |
| Console: | Bruker Avance III |
| Probes: | Triple Res. Cryo-TCI (1H/13C/15N) Broadband Inverse BBI (1H, BB) Z-Gradient, ATM |
| Automation: | BACS 60 |
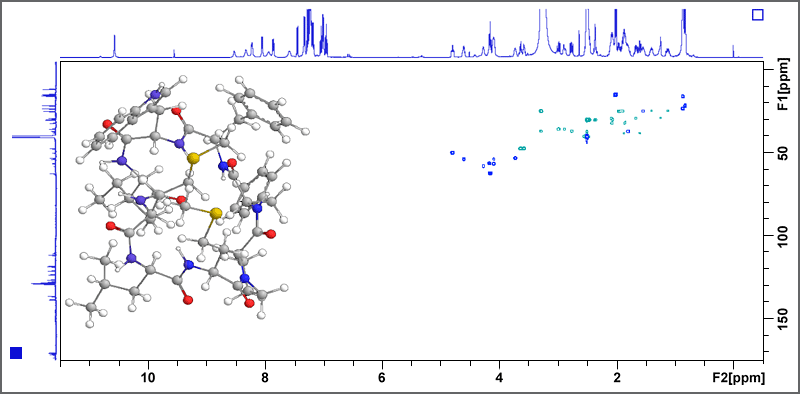
The 500 MHz AV III system is used for the structure determination of previously unknown bioactive natural products, functional food constituents, and metabolites. COSY, TOCSY, HSQC, HMBC, ADEQUATE, and INADEQUATE experiments give valuable information on the constitution of the target molecule, whereas NOESY and ROESY experiments deliver important insights into the structural conformation of the molecule. Fig. 1 displays the example of a HSQC spectrum of a bitter tasting cyclic octapeptide isolated from linseed oil.
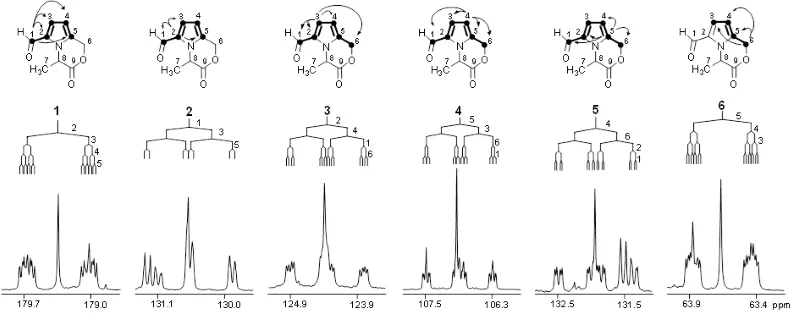
In order to trace chemical transformation pathways and perform metabolic flux analysis, stable isotope labelling experiments are performed with NMR diagnostic analysis of 13C-13C couplings (e.g. the carbon bond labelling technique CABOLA; Fig. 2) to visualize the time-resolved joint transfer of labelled atoms from a precursor into a target molecule.
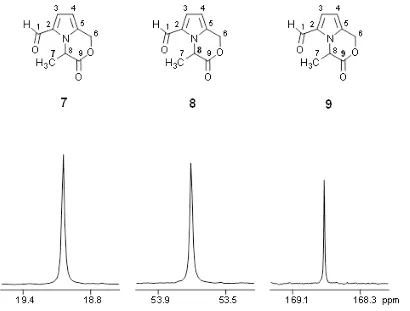
For purity assessment, as well as target compound quantitation, quantitative NMR (qNMR) spectroscopy is performed on the NMR system (qHNMR, Fig. 3, and q13C-NMR). Using the ERETIC2 software tool, quantification is performed using an external calibrant within an error limit of ± 2%. Quantitative analysis of target molecules in complex natural compound mixtures, such as e.g. biosamples (urine, plasma etc.), food (coffee, wine etc.), or fermentation broths is performed by 1H-NMR spectroscopy with or without water pre-saturation and in addition with J-resolved experiments.
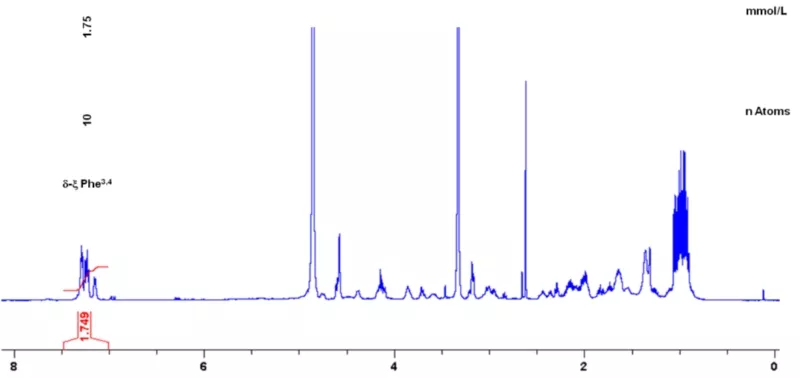
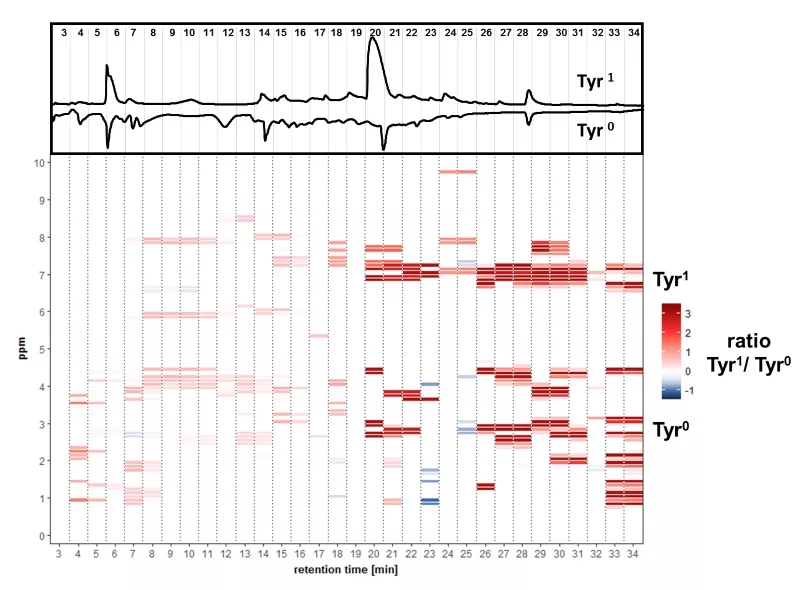
A differential off-line LC-NMR approach (DOLC-NMR) has been developed to capture and quantitate nutrient-induced metabolome alterations e.g. in microorganisms like Saccharomyces cerevisiae. Off-line coupling of HPLC separation and 1H-NMR spectroscopy supported by automated comparative bucket analyses, followed by quantitative 1H-NMR using ERETIC 2 allows the visualization of quantitative changes in the strain's metabolome. As an example, the DOLC-NMR spectrum showing metabolome alterations in S. cerevisiae induced by the amino acid L-tyrosine is displayed in Fig. 4.