P2: Genetic basis and gene regulation of root trait driven beneficial root-microbe interaction
Beneficial interactions with microorganisms are pivotal for crop performance and resilience. However, it remains unclear how heritable the microbiome is with respect to the host plant genotype and to what extent host genetic mechanisms can modulate plant–microbiota interactions in the face of environmental stresses.
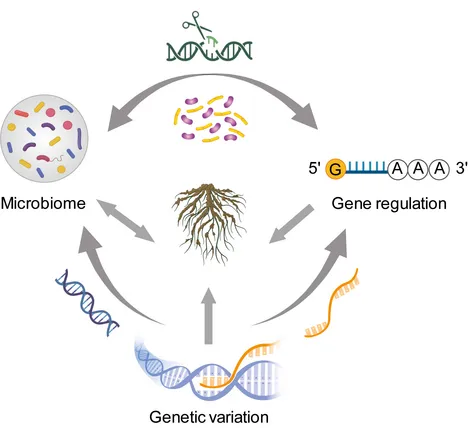
This project enables the deep understanding of reciprocal interactions between host plants and rhizosphere microbes via gene expression and metabolic signature, meanwhile we consider the plant phenotype (i.e. root and rhizosphere trait) is the central component in mediating such beneficial interactions. Several novel approaches combined with plant genome editing, high-throughput bacteria cultivation and cell specific analyses are ongoing.
Key publication:
- Yu P, He X, Baer M, Beirinckx S, Tian T, Moya YAT, Zhang Y, Deichmann M, Frey FP, Bresgen V, Li C, Razavi BS, Schaaf G, von Wirén N, Su Z, Bucher M, Tsuda K, Goormachtig S, Chen X, Hochholdinger F "Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation." Nature Plants. 2021; 7: 481–499.
- Wang D, He X, Baer M, Lami K, Yu B, Tassinari A, Salvi S, Schaaf G, Hochholdinger F, Yu P "Lateral root enriched Massilia associated with plant flowering in maize." Microbiome. 2024; 12(1): 124.