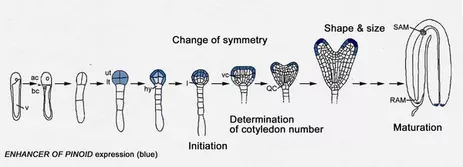
Plant embryogenesis is marked by several critical stages. One key event is the elaboration of the bilateral symmetry from a formely radial symmetry. In dicots globular stage embryos are characterised by a radial symmetry and do not exhibit cotyledon primordia. During transition stage the embryo developes two cotyledons leading to the bilateral symmetry of the heart stage embryo.
We have disected the development of cotyledons from the initiation to the final growth stages genetically such that at least three steps can be discriminated: 1. initiation, 2. determination of cotyledon number and 3. coordinated growth to reach normal size and shape.
GENES CONTROLLING COTYLEDON NUMBER AND DEVELOPMENT

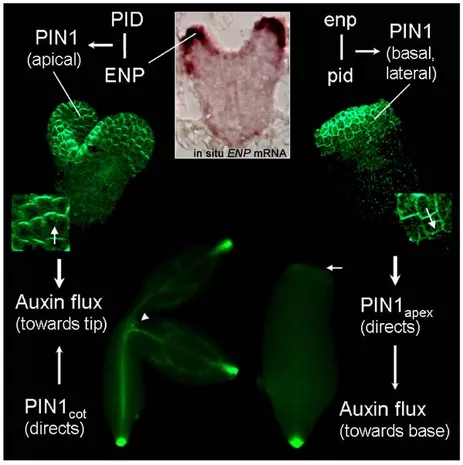
The most advanced study in our lab focusses on mutants which fail to initiate cotyledons. Genes responsible for cotyledon initiation are now identified (Treml et al., 2005; Furutani et al., 2007). These genes interfere with the asymmetric distribution of the hormon auxin, which is required for the initiation and maintenance of plant organs. These local auxin concentrations (so-called auxin maxima) also emerge in the tips of cotyledons during embryogenesis. The transport/distribution of auxin is carried out by the integral membrane PIN1 transporter whose polar localization is controlled by Ser/Thr-kinase PINOID (PID). Seedlings of single mutants of PID exhibit irregular cotyledon numbers, but the cotyledons look normal. Our lab could show that the simultaneous action of PID and ENHANCER OF PINOID (ENP) controls the initiation of cotyledon primordia (Treml et al., 2005). This analysis showed that in pid single mutants most PIN1 molecules are polarly localized. However, simultaneous mutation of ENP and PID completely abolishes apical PIN1 localization in the epidermis of embryos. Instead PIN1 adopts a basal polarity (its default state) and any auxin synthesized in these cells will be transported downwards towards the root tip. As a consequence, auxin maxima are not established at the presumptive cotyledon primordia tips. This in turn leads to cotyledon-less embryos. These observations are integrated into our current model of cotyledon initiation.

We have recently summarized the most important insights into PID and ENP controlled PIN1 polarity and its significance for cotyledon primordia initiation in Matthes and Torres-Ruiz (2012) Initiation und Etablierung von keimblättern im Arabidopsis-Embryo. BIOspektrum 07, 717-720.
The predominant effect of a second group of apical mutants is the alteration of cotyledon numbers, which leads to seedlings with one, three or even more cotyledons (unpublished work). mon1 and rpk1-7 (formerly FN9-3) mutants affect cotyledon number and possess an intact hypocotyl region and root. We have recently analyzed the effects of rpk1-7 and rpk1-6 in more detail (Luichtl et al., 2013). Apparently, epidermal cell shape and polarity are compromised in rpk1 embryos, as evidenced by disturbed polarity of the auxin efflux carrier PIN1. The effects of rpk1 manifest in a spatially and timely stochastic fashion. This is probably caused by redundancy of RPK1-like functions encoded in the Arabidopsis genome. As a corollary of PIN1 disturbance, auxin maxima show a stochastic distribution in rpk1 embryos, being at times entirely absent and at other times supernumerary. We have developed a model which accounts for this variability in order to explain how monocotyledonous seedlings and cotyledon shape variants can developmentally arise in Arabidopsis and possibly in other plants (Luichtl et al., 2013).
From this and other work we are also learning that, in concert with the genes mentioned above, modifier genes present in different genetic backgrounds have significant influence in the decision, which leads to the development of a cotyledon. For detecting the presence of such modifiers, we have used Arabidopsis ecotypes whose genetic variability had been demonstrated in an earlier work of our lab (Erschadi et al., 2000). Note that the work of Yu et al. (2009, 2010) on tetraploid Arabidopsis ecotypes is also related to genetic diversity and natural variation (see below).
Further reading:
Luichtl M, Fiesselmann BS, Matthes M, Yang X, Peis O, Brunner A, Torres Ruiz RA (2013) Mutations in the Arabidopsis RPK1 gene uncouple cotyledon anlagen and primordia by modulating epidermal cell shape and polarity. Biology Open, DOI: 10.1242/bio.20135991 (Open Access)
Furutani, M.; Kajiwara, T.; Kato, T.; Treml, B.S.; Stockum, C.; Torres-Ruiz, R.A. and Tasaka, M. (2007) The gene MACCHI-BOU4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism on the molecular level. Development 134, 3849-3859.
Treml, B. S.; Winderl, S.; Radykewicz, R.; Herz, M., Schweizer, G., Hutzler, P., Glawischnig, E; Torres Ruiz, R. A. (2005) The gene ENHACER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development, 132: 4063-4074.
see also commentary: "Reversed auxin transport jams development" Development 2005 132:e1802
Torres-Ruiz, R. A. (2004) "Polarity in Arabidopsis embryogenesis" in "Polarity in plants" (K. Lindsey ed.) Annual Plant Reviews Vol.12. Blackwell Publishing, Oxford, pp 223-261(review on different aspects of embryo development)
Geldner, N., Richter, S., Vieten, A., Marquardt, S., Torres-Ruiz, R. A., Mayer, U. and Jürgens, G. (2004). Partial loss-of-function alleles reveal a role for GNOM in post-embryonic and auxin-transport related development of Arabidopsis. Development 131, 389-400.(shows what is not a cotyledon-specific gene but in this special case looks as if, i. e. weak alleles of GNOM result in monocotyledoneous mutants).
Erschadi, S.; Haberer, G.; Schöniger, M. and Torres Ruiz, R. A. (2000) Estimating genetic diversity of Arabidopsis thaliana ecotypes with Amplified Fragment Length Polymorphisms (AFLP).Theor Appl Genet 100, 633-640.
ANALYSIS OF APICAL PATTERN AND PROLIFERATION GENES OF ARABIDOPSIS THALIANA
We have isolated mutants in which the apical meristem proliferates abnormally, producing numerous aberrant leaves and shoots, an effect reminiscent of Cytokinin-Auxin imbalance. When cultured at elevated cytokinin concentrations the mutants respond with enhanced proliferation (unpublished data). Mutants of this type represent genes like GURKE and PEPINO (Mayer et al., 1991; TorresRuiz et al., 1996a, Torres Ruiz et al., 1996b). A further mutant which belongs to this group, pasticcino1, has been characterised and the corresponding gene has recently been cloned (Faure et al., 1998, Development 125,pp 909-918; Vittorioso et al. 1998, Mol Cell Biol 18, pp 3034-3043). Our molecular work has been or is focussed on the positional cloning of these genes since transposon- or T-DNA-tagging populations did not provide corresponding mutants. We have established different mapping techniques in our lab, isolated closely linked molecular markers and established contigs to narrow down the physical regions covering these genes (Haberer et al., 1996; Torres Ruiz et al., 1996a, unpublished data). PEPINO has been cloned in our lab. We have sequenced wild-type and mutant alleles of this gene (see below) and now aim to a more detailed molecular analysis in order to elucidate its function.
Further reading:
Sabine Schönhofer-Merl and Ramón A. Torres-Ruiz (2010) The Sos-recruitment system as a tool to analyze cellular localization of plant proteins: membrane localization of Arabidopsis thaliana PEPINO/PASTICCINO2. Mol Genet Genomics 283: 439-449.
Haberer, G., Erschadi, S. and Torres-Ruiz, R. A. (2002) The Arabidopsis gene PEPINO/PASTICCINO2 is required for proliferation control of meristematic and non-meristematic cells and encodes a putative anti-phosphatase. Dev Genes and Evol 212, 542-550.
ANALYSIS OF MORPHOGENESIS GENES OF ARABIDOPSIS THALIANA

The work in this area concentrated on the characterisation of these mutants and the identification of new ones in addition to those mutants previously found (fass, knopf and mickey; Mayer et al., 1991 see above). The developmental, genetic and histological analysis of these mutants points to different important events during the elaboration of the morphogenesis of cells (and in turn of the plant body). In fass mutants the adequate orientation of newly formed cell walls is disturbed from the first zygotic division onwards (Torres Ruiz and Jürgens, 1994). The resulting seedlings exhibit an extreme compression of all organs along the apical-basal and proximo-distal axes of the plant. fass plants do not produce preprophase bands (Traas et al. 1995, Nature 375, pp 676-677), cytoskeletal structures consisting of microtubules which mark the future plane of cell division in dividing cells. Mutations in the genes KNOPF and ZWERGERL lead as well to small miniaturised seedlings. However, in contrast to fass, it seems that in these mutants the process promoting apical-basal elongation of cells is broken down. As a result especially epidermal and cortical cells of the hypocotyl region expand abnormally along the radial axis of the embryo (Torres Ruiz et al., 1996a and unpublished data).
ANALYSIS OF POLYPLOIDY IN PLANTS
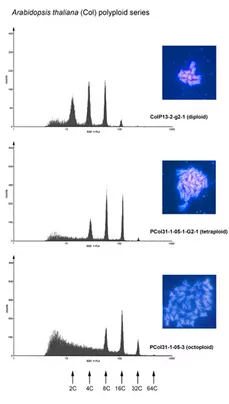
Polyploidy is a widespread phenomenon in animals and plants. Possibly all angiosperm genomes (except that of the basal species Amborella) bear traces of polyploidization. Two forms of polyploidy are frequently considered allopolyploidy, which results from interspecies hybridization and autopolyploidy, which originates from genome duplication within one particular species. However, reports indicate that there is a continuum of polyploid situations between these extremes (e. g. Soltis and Soltis, Annu Rev Plant Biol 2009, Vol. 60, pp. 561-588). Polyploidy, in particular tetraploidy, has several potential advantages probably because the organisms can resort to a higher number of genes and a higher maximum number of allelic variants. This is believed to be advantageous for plant metabolism in terms of elevated rates of synthesis or a higher variability of metabolically relevant compounds. A further advantage lies in the subfunctionalization of gene copies, that take over the task in different tissues. Plant breeders have taken advantage of polyploidy in order to improve agronomic traits of economically important plants some of which have been generated by allopolyploidisation of interspecies hybrids or by autopolyploidisation within species.
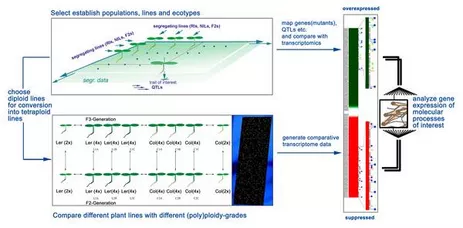
An advanced molecular understanding of plant polyploidy will help to understand this phenomenon, which is of crucial importance for plant breeding and evolution. Arabidopsis thaliana has, in conjunction with A. arenosa, developed into a system for the molecular analysis of alloplolyploidy. However, there are only few Arabidopsis lines available to study autopolyploidy. In order to be able to investigate polyploidy on a reliable basis, we have optimised conventional methodologies and developed a novel strategy for the rapid generation and identification of polyploids based on trichome branching patterns (Yu et al., 2009). The analysis of over two dozens independently induced Arabidopsis lines has lead to interesting observations concerning the relationship between cell size and ploidy levels and on the relative stability of tetraploidy in Arabidopsis over at least three consecutive generations. The most important finding of this work is that neo-tetraploid lines exhibit considerable stability through all the generations tested. The systematic generation of tetraploid collections through this strategy as well as the lines generated in this work will help to unravel the consequences of polyploidy, in particular tetraploidy, on the genome, on gene expression and on natural diversity in Arabidopsis. The establishment of these lines will enable us to analyze autotetraploidy in more detail. We are currently analyzing the transcriptome of these lines by means of whole genome DNA-microarray analysis. The bioinformatic evaluation of these data is carried out in collaboration with the groups of KFX Mayer (Helmholtz-Inst, München) and T. Rattei (Univ. Wien). We have learned that the impact on the transcriptome is different between A. thaliana tetraploid ecotypes (Yu et al., 2010).
The generation of polyploid lines not only leads to plants with multiples of regular chromosome sets but also to aberrant chromosome sets and aberrant chromosomes. We are therefore interested in studying the structure of chromosomes and in particular the centromere region. In the process of positional cloning of the GURKE gene we learned about the structure and perculiarities of this important chromosome element. In a collaboration with Dr. P. Fransz (University of Wageningen) this work has provided interesting genetic, cytological and molecular data, which have been partly published (Fransz et al., 1998; W. Haupt et al. 2001). Many new unknown repetitive elements have been found, one group, which flanks all Arabidopsis centromeres is of particular interest. We are investigating the significance of these elements as well as that of others in context of essential functions of the centromere. The data obtained so far point to a plant centromere structure, which is discussed as being similar to that of animals and humans (Hemleben et al., 2000). Increased knowledge on the centromere, its structural analysis and the localisation of functional regions might, as long term goal, allow the construction of plant artificial minichromosomes (PLACs).
Further reading:
Yu Z, Haage K, Streit VE, Gierl A, Torres-Ruiz RA (2009) A large number of tetraploid Arabidopsis thaliana lines, generated by a rapid strategy, reveal high stability of neo-tetraploids during consecutive generations. Theor Appl Genet 118, 1107-1119.
Zheng Yu, Georg Haberer, Michaela Matthes, Thomas Rattei, Klaus F. X. Mayer, Alfons Gierl and Ramon A. Torres-Ruiz (2010) Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc Nat Acad Sci USA 107, 17809-17814.
Haupt, W.; Fischer, T.C.; Winderl, S.; Fransz, P. and Torres Ruiz, R. A. (2001). The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. The Plant J 27, 285-296.