J. D. Berndt and W. Wong, 2014: Signaling Breakthroughs of the Year. Sci. Signal. 8, eg1 (2015).
Citation from the full text:
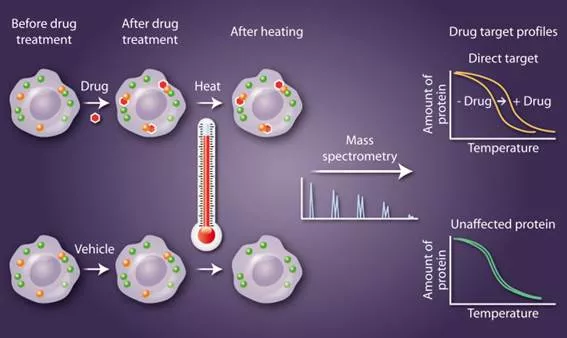
"Every year, we receive nominations for breakthrough methods that address difficult biological questions. New technologies not only facilitate research but also provide data that lead scientists to pose questions that were not previously evident. Cancer drug discovery often begins by screening large libraries of small molecules for the ability to kill cultured cancer cells. One bottleneck in the development pipeline is the identification of the molecular target or targets of these molecules. Janes suggested a study by Savitski et al. (33) that combines quantitative mass spectrometry analysis of cell lysates with ligand-induced changes in protein thermal stability to identify protein-drug interactions (Fig. 5). Janes wrote, “This paper introduces a remarkably simple method for monitoring protein conformational changes triggered by small molecules, such as kinase inhibitors and second messengers. The technique provides information about signaling networks that is complementary to that obtained by other proteomic methods. One of its appeals is that thermal stability profiling can be implemented in an unbiased way by using isobaric tags and mass spectrometry, or it can be done in a focused way by immunoblotting for specific target proteins. I found it especially striking that the technique identified a heme biosynthesis enzyme [FECH] as an off-target of protein kinase inhibitors with recognized phototoxicity side effects in the clinic. I would be eager to see thermal proteome profiling applied systematically across all FDA-approved kinase inhibitors.” Moreover, genetic defects in FECH are associated with photosensitivity in humans, suggesting that drug-mediated inhibition of FECH may be a mechanism for this common side effect."